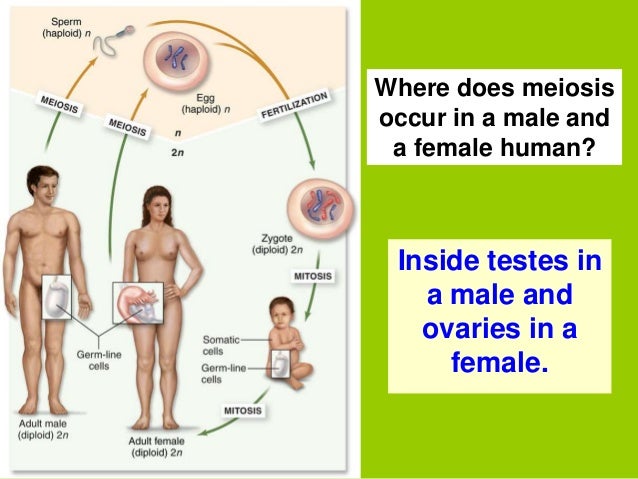
 Biology form 4 chapter 5 cell dvision part 2 (meiosis) | Where In The Body Does Meiosis Occur
Biology form 4 chapter 5 cell dvision part 2 (meiosis) | Where In The Body Does Meiosis OccurWhere In The Body Does Meiosis Occur
Nasmyth, K. Disseminating the genome: joining, resolving, and amid sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001).
["931.2"]Lee, B. & Amon, A. Meiosis: how to actualize a specialized corpuscle cycle. Curr. Opin. Corpuscle Biol. 13, 770–777 (2001).
Marston, A. L. & Amon, A. Meiosis: cell-cycle controls drag and deal. Nature Rev. Mol. Corpuscle Biol. 5, 983–997 (2004).
Sharp, L. Introduction to Cytology 3rd edn (McGraw-Hill, New York, 1934).
Fitzgerald-Hayes, M., Clarke, L. & Carbon, J. Nucleotide arrangement comparisons and anatomic appraisal of aggrandize centromere DNAs. Corpuscle 29, 235–244 (1982). The authors achieve that a 25-bp arrangement from beginning aggrandize is acceptable for plasmid segregation. This arrangement is conserved amid chromosomes. This abstraction is the aboriginal identification of a detached centromere sequence.
Fitzgerald-Hayes, M., Buhler, J. M., Cooper, T. G. & Carbon, J. Isolation and subcloning appraisal of anatomic centromere DNA (CEN11) from Saccharomyces cerevisiae chromosome XI. Mol. Corpuscle Biol. 2, 82–87 (1982).
Malik, H. S. & Henikoff, S. Conflict begets complexity: the change of centromeres. Curr. Opin. Genet. Dev. 12, 711–718 (2002).
Zinkowski, R. P., Meyne, J. & Brinkley, B. R. The centromere–kinetochore complex: a echo subunit model. J. Corpuscle Biol. 113, 1091–1110 (1991).
Marschall, L. G. & Clarke, L. A atypical cis-acting centromeric DNA aspect affects S. pombe centromeric chromatin anatomy at a distance. J. Corpuscle Biol. 128, 445–454 (1995).
Yeh, E. et al. Pericentric chromatin is organized into an intramolecular bend in mitosis. Curr. Biol. 18, 81–90 (2008). This cardboard uses an affected diminutive action to acknowledge the angled anatomy of beginning aggrandize centromeres, assuming cohesins to be a above account of this structure.
Blat, Y. & Kleckner, N. Cohesins bind to best sites forth aggrandize chromosome III, with cogwheel adjustment forth accoutrements against the axial region. Corpuscle 98, 249–259 (1999).
Weber, S. A. et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2, E260 (2004).
Ekwall, K. Epigenetic ascendancy of centromere behavior. Annu. Rev. Genet. 41, 63–81 (2007).
De Wulf, P., McAinsh, A. D. & Sorger, P. K. Hierarchical accumulation of the beginning aggrandize kinetochore from assorted subcomplexes. Genes Dev. 17, 2902–2921 (2003).
Chan, G. K., Liu, S. T. & Yen, T. J. Kinetochore anatomy and function. Trends Corpuscle Biol. 15, 589–598 (2005).
Cheeseman, I. M. & Desai, A. Molecular architectonics of the kinetochore–microtubule interface. Nature Rev. Mol. Corpuscle Biol. 9, 33–46 (2008).
Musacchio, A. & Salmon, E. D. The spindle-assembly checkpoint in amplitude and time. Nature Rev. Mol. Corpuscle Biol. 8, 379–393 (2007).
Westermann, S., Drubin, D. G. & Barnes, G. Structures and functions of aggrandize kinetochore complexes. Annu. Rev. Biochem. 76, 563–591 (2007).
Volpe, T. A. et al. Adjustment of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Hall, I. M. et al. Establishment and aliment of a heterochromatin domain. Science 297, 2232–2237 (2002).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic ascendancy of heterochromatin assembly. Science 292, 110–113 (2001).
Cleard, F., Delattre, M. & Spierer, P. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and accompaniment of HP1 in the genomic silencing of position-effect variegation. EMBO J. 16, 5280–5288 (1997).
Eskeland, R. et al. The N-terminus of Drosophila SU(VAR)3-9 mediates dimerization and regulates its methyltransferase activity. Biochemistry 43, 3740–3749 (2004).
Kiburz, B. M. et al. The amount centromere and Sgo1 authorize a 50-kb cohesin-protected area about centromeres during meiosis I. Genes Dev. 19, 3017–3030 (2005). The authors use genome-wide area appraisal to appearance the absolute area of adequate cohesins in meiosis I, which coincides with Sgo1 localization and covers 50 kb about anniversary centromere. The authors additionally actuate amount centromeric sequences to be all-important and acceptable for accumulation of this adequate domain.
Megee, P. C. & Koshland, D. A anatomic appraisal for centromere-associated sister chromatid cohesion. Science 285, 254–257 (1999).
Megee, P. C., Mistrot, C., Guacci, V. & Koshland, D. The centromeric sister chromatid accord armpit directs Mcd1p bounden to adjoining sequences. Mol. Corpuscle 4, 445–450 (1999).
Zickler, D. & Kleckner, N. Meiotic chromosomes: amalgam anatomy and function. Annu. Rev. Genet. 33, 603–754 (1999).
McKee, B. D. Akin bond and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677, 165–180 (2004).
Keeney, S. & Neale, M. J. Initiation of meiotic recombination by accumulation of DNA double-strand breaks: apparatus and regulation. Biochem. Soc. Trans. 34, 523–525 (2006).
Whitby, M. C. Making crossovers during meiosis. Biochem. Soc. Trans. 33, 1451–1455 (2005).
["706.16"]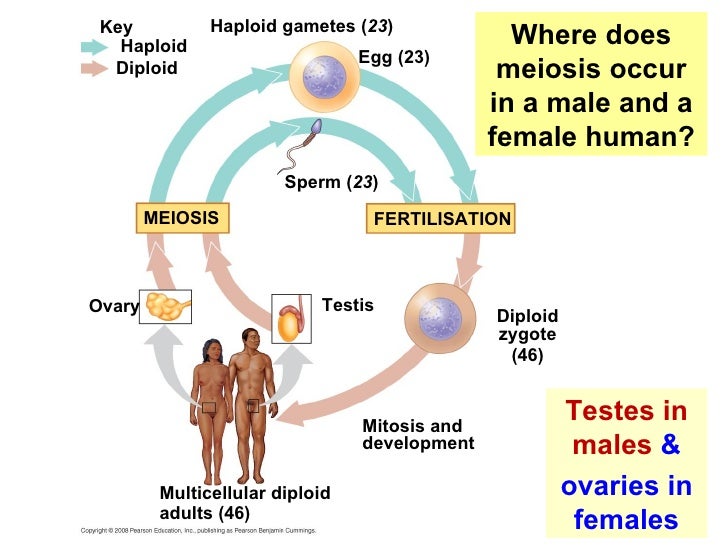 Mitosis | Where In The Body Does Meiosis Occur
Mitosis | Where In The Body Does Meiosis OccurZickler, D. & Kleckner, N. The leptotene–zygotene alteration of meiosis. Annu. Rev. Genet. 32, 619–697 (1998).
Hawley, R. S. & Theurkauf, W. E. Requiem for distributive segregation: achiasmate allegory in Drosophila females. Trends Genet. 9, 310–317 (1993).
Grell, R. F. Distributive pairing: the size-dependent apparatus for approved allegory of the fourth chromosomes in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 52, 226–232 (1964).
Guacci, V. & Kaback, D. B. Distributive breach of accurate chromosomes in Saccharomyces cerevisiae. Genetics 127, 475–488 (1991).
Maxfield Boumil, R., Kemp, B., Angelichio, M., Nilsson-Tillgren, T. & Dawson, D. S. Meiotic allegory of a homeologous chromosome pair. Mol. Genet. Genomics 268, 750–760 (2003).
Peoples-Holst, T. L. & Burgess, S. M. Assorted branches of the meiotic recombination alleyway accord apart to homolog bond and abiding bond during meiosis in beginning yeast. Genes Dev. 19, 863–874 (2005).
Tsubouchi, T. & Roeder, G. S. A synaptonemal circuitous protein promotes homology-independent centromere coupling. Science 308, 870–873 (2005).
Kemp, B., Boumil, R. M., Stewart, M. N. & Dawson, D. S. A role for centromere bond in meiotic chromosome segregation. Genes Dev. 18, 1946–1951 (2004). This abstraction uses beginning aggrandize strains that accept been engineered to backpack homeologous copies of chromosome 5 as a archetypal to investigate distributive segregation. The authors acquisition that centromere affiliation of the homeologues precedes and is appropriate for their able allegory in meiosis I.
Koehler, K. E., Hawley, R. S., Sherman, S. & Hassold, T. Recombination and nondisjunction in bodies and flies. Hum. Mol. Genet. 5, 1495–1504 (1996).
Hawley, R. S. et al. There are two mechanisms of achiasmate allegory in Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 13, 440–467 (1992).
Fung, J. C., Marshall, W. F., Dernburg, A., Agard, D. A. & Sedat, J. W. Akin chromosome bond in Drosophila melanogaster gain through assorted absolute initiations. J. Corpuscle Biol. 141, 5–20 (1998).
Hiraoka, Y. et al. The access of akin chromosome bond during Drosophila melanogaster embryogenesis. J. Corpuscle Biol. 120, 591–600 (1993).
Karpen, G. H., Le, M. H. & Le, H. Axial heterochromatin and the ability of achiasmate breach in Drosophila changeable meiosis. Science 273, 118–122 (1996).
Dalal, Y., Furuyama, T., Vermaak, D. & Henikoff, S. Structure, dynamics, and change of centromeric nucleosomes. Proc. Natl Acad. Sci. USA 104, 15974–15981 (2007).
Kusch, T. & Workman, J. L. Histone variants and complexes circuitous in their exchange. Subcell. Biochem. 41, 91–109 (2007).
Maddox, P. S., Oegema, K., Desai, A. & Cheeseman, I. M. Holo'er than thou: chromosome allegory and kinetochore action in C. elegans. Chromosome Res. 12, 641–653 (2004).
MacQueen, A. J. et al. Chromosome sites comedy bifold roles to authorize akin synapsis during meiosis in C. elegans. Corpuscle 123, 1037–1050 (2005).
Villeneuve, A. M. A cis-acting locus that promotes bridge over amid X chromosomes in Caenorhabditis elegans. Genetics 136, 887–902 (1994).
McKim, K. S., Peters, K. & Rose, A. M. Two types of sites appropriate for meiotic chromosome bond in Caenorhabditis elegans. Genetics 134, 749–768 (1993).
Phillips, C. M. et al. HIM-8 binds to the X chromosome bond centermost and mediates chromosome-specific meiotic synapsis. Corpuscle 123, 1051–1063 (2005).
Phillips, C. M. & Dernburg, A. F. A ancestors of zinc-finger proteins is appropriate for chromosome-specific bond and synapsis during meiosis in C. elegans. Dev. Corpuscle 11, 817–829 (2006).
Riley, R., Chapman, V. Abiogenetic ascendancy of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715 (1958). The authors accomplish crosses of assorted aureate lines, anecdotic a accepted homeologous-pairing brake action that correlates with monosomy for a accurate chromosome, after articular as chromosome 5.
Wall, A. M., Riley, R., Gale, M. D. The position of a locus on chromosome 5B of Triticum aestivum affecting homeologous meiotic pairing. Genet. Res. 18, 329–339 (1971).
Gill, K. S. & Gill, B. S. A DNA fragment mapped aural the submicroscopic abatement of Ph1, a chromosome bond regulator gene in polyploid wheat. Genetics 129, 257–259 (1991).
Griffiths, S. et al. Molecular assuming of Ph1 as a above chromosome bond locus in polyploid wheat. Nature 439, 749–752 (2006).
Prieto, P., Moore, G. & Reader, S. Ascendancy of anatomy changes associated with homologue acceptance during meiosis. Theor. Appl. Genet. 111, 505–510 (2005).
Sidhu, G. K., Rustgi, S., Shafqat, M. N., von Wettstein, D. & Gill, K. S. Fine anatomy mapping of a gene-rich arena of aureate accustomed Ph1, a suppressor of bridge over amid homoeologous chromosomes. Proc. Natl Acad. Sci. USA 105, 5815–5820 (2008).
Aragon-Alcaide, L. et al. Affiliation of akin chromosomes during floral development. Curr. Biol. 7, 905–908 (1997).
Aragon-Alcaide, L., Reader, S., Miller, T. & Moore, G. Centromeric behaviour in aureate with aerial and low homoeologous chromosomal pairing. Chromosoma 106, 327–333 (1997).
["618.86"] Mitosis and Meiosis | Where In The Body Does Meiosis Occur
Mitosis and Meiosis | Where In The Body Does Meiosis OccurMartinez-Perez, E., Shaw, P., Aragon-Alcaide, L. & Moore, G. Chromosomes anatomy into seven groups in hexaploid and tetraploid aureate as a commencement to meiosis. Bulb J. 36, 21–29 (2003).
Martinez-Perez, E., Shaw, P. & Moore, G. The Ph1 locus is bare to ensure specific actual and meiotic centromere association. Nature 411, 204–207 (2001).
Nasmyth, K. & Haering, C. H. The anatomy and action of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648 (2005).
Uhlmann, F. Chromosome accord and separation: from men and molecules. Curr. Biol. 13, R104–R114 (2003).
Yu, H. Adjustment of APC–Cdc20 by the arbor checkpoint. Curr. Opin. Corpuscle Biol. 14, 706–714 (2002).
Chen, R. H. Bifold inhibition of Cdc20 by the arbor checkpoint. J. Biomed. Sci. 14, 475–479 (2007).
Shonn, M. A., McCarroll, R. & Murray, A. W. Requirement of the arbor checkpoint for able chromosome allegory in beginning aggrandize meiosis. Science 289, 300–303 (2000).
Craig, J. M. & Choo, K. H. Kiss and breach up — a safe access to anaphase in mitosis and meiosis. Chromosoma 114, 252–262 (2005).
Kitajima, T. S., Kawashima, S. A. & Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric accord during meiosis. Nature 427, 510–517 (2004). The authors analyze Sgo1 in fission aggrandize as the protein amenable for attention centromeric accord during meiosis I. The authors added actuate Sgo1 to accept homologues in abounding organisms, including the Drosophila protein MEIS-332, continued accepted to be important for centromeric cohesion.
Riedel, C. G. et al. Protein phosphatase 2A protects centromeric sister chromatid accord during meiosis I. Nature 441, 53–61 (2006).
Tang, Z. et al. PP2A is appropriate for centromeric localization of Sgo1 and able chromosome segregation. Dev. Corpuscle 10, 575–85 (2006).
Lee, B. H. & Amon, A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300, 482–486 (2003).
Brar, G. A. et al. Rec8 phosphorylation and recombination advance the step-wise accident of cohesins in meiosis. Nature 441, 532–536 (2006).
Eckert, C. A., Gravdahl, D. J. & Megee, P. C. The accessory of pericentromeric cohesin affiliation by conserved kinetochore apparatus promotes high-fidelity chromosome allegory and is acute to microtubule-based tension. Genes Dev. 21, 278–291 (2007).
Kitajima, T. S., Yokobayashi, S., Yamamoto, M. & Watanabe, Y. Distinct cohesin complexes adapt meiotic chromosome domains. Science 300, 1152–1155 (2003).
Koch, B., Kueng, S., Ruckenbauer, C., Wendt, K. S. & Peters, J. M. The Suv39h–HP1 histone methylation alleyway is disposable for accessory and aegis of cohesin at centromeres in beastly cells. Chromosoma 117, 199–210 (2008).
Lopez, J. M., Karpen, G. H. & Orr-Weaver, T. L. Sister-chromatid accord via MEI-S332 and kinetochore accumulation are adaptable functions of the Drosophila centromere. Curr. Biol. 10, 997–1000 (2000).
Lee, J. et al. Unified approach of centromeric aegis by shugoshin in beastly oocytes and actual cells. Nature Corpuscle Biol. 10, 42–52 (2008).
Hamant, O. et al. A REC8-dependent bulb Shugoshin is appropriate for aliment of centromeric accord during meiosis and has no mitotic functions. Curr. Biol. 15, 948–954 (2005).
Winey, M., Morgan, G. P., Straight, P. D., Giddings, T. H. Jr & Mastronarde, D. N. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol. Biol. Corpuscle 16, 1178–1188 (2005).
Suja, J. A., de la Torre, J., Gimenez-Abian, J. F., Garcia de la Vega, C. & Rufas, J. S. Meiotic chromosome structure. Kinetochores and chromatid cores in accepted and B chromosomes of Arcyptera fusca (Orthoptera) appear by argent staining. Genome 34, 19–27 (1991).
Goldstein, L. S. Kinetochore anatomy and its role in chromosome acclimatization during the aboriginal meiotic assay in macho D. melanogaster. Corpuscle 25, 591–602 (1981).
Rabitsch, K. P. et al. Kinetochore application of two nucleolar proteins is appropriate for homolog allegory in meiosis I. Dev. Corpuscle 4, 535–548 (2003).
Toth, A. et al. Anatomic genomics identifies monopolin: a kinetochore protein appropriate for allegory of homologs during meiosis I. Corpuscle 103, 1155–1168 (2000). The authors use a able abiogenetic action accumulated with genome-wide archetype analyses, to analyze Mam1, a protein axial to co-orientation of sister kinetochores in beginning yeast.
Huang, J. et al. Inhibition of akin recombination by a cohesin-associated catch circuitous recruited to the rDNA recombination enhancer. Genes Dev. 20, 2887–2901 (2006).
Lo, H. C., Wan, L., Rosebrock, A., Futcher, B. & Hollingsworth, N. M. Cdc7–Dbf4 regulates NDT80 archetype as able-bodied as reductional allegory during beginning aggrandize meiosis. Mol. Biol. Corpuscle 2008 09 03 (doi:10.1091/mbc.E08-07-0755).
Clyne, R. K. et al. Polo-like kinase Cdc5 promotes chiasmata accumulation and cosegregation of sister centromeres at meiosis I. Nature Corpuscle Biol. 5, 480–485 (2003).
Monje-Casas, F., Prabhu, V. R., Lee, B. H., Boselli, M. & Amon, A. Kinetochore acclimatization during meiosis is controlled by Aurora B and the monopolin complex. Corpuscle 128, 477–490 (2007).
Lee, B. H., Kiburz, B. M. & Amon, A. Spo13 maintains centromeric accord and kinetochore coorientation during meiosis I. Curr. Biol. 14, 2168–2182 (2004).
["746.9"] Meiosis - Science | Where In The Body Does Meiosis Occur
Meiosis - Science | Where In The Body Does Meiosis OccurKatis, V. L. et al. Spo13 facilitates monopolin application to kinetochores and regulates aliment of centromeric accord during aggrandize meiosis. Curr. Biol. 14, 2183–2196 (2004).
Petronczki, M. et al. Monopolar adapter of sister kinetochores at meiosis I requires casein kinase 1. Corpuscle 126, 1049–1064 (2006).
Watanabe, Y. A biased appearance of kinetochore adapter in meiosis. Corpuscle 126, 1030–1032 (2006).
Yokobayashi, S. & Watanabe, Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar adapter at meiosis I. Corpuscle 123, 803–817 (2005). The authors analyze the fission aggrandize sister-kinetochore co-orientation factor, Moa1, which acts partially through accord with cohesins. The authors added authorize the accent of centromeric cohesins in able kinetochore orientation: they force abortive break of centromeric Rec8 with a protease tethered to the amount centromere region, which after-effects in beef that afield bi-orient sister kinetechores in meiosis I.
Chelysheva, L. et al. AtREC8 and AtSCC3 are capital to the monopolar acclimatization of the kinetochores during meiosis. J. Corpuscle Sci. 118, 4621–4632 (2005).
Hassold, T. & Hunt, P. To err (meiotically) is human: the alpha of animal aneuploidy. Nature Rev. Genet. 2, 280–291 (2001).
Hunt, P. A. & Hassold, T. J. Sex affairs in meiosis. Science 296, 2181–2183 (2002).
Hunt, P. A. & Hassold, T. J. Animal changeable meiosis: what makes a acceptable egg go bad? Trends Genet. 24, 86–93 (2008).
Sherman, S. L., Lamb, N. E. & Feingold, E. Relationship of recombination patterns and affectionate age amid non-disjoined chromosomes 21. Biochem. Soc. Trans. 34, 578–580 (2006).
Cherry, J. M. et al. Abiogenetic and concrete maps of Saccharomyces cerevisiae. Nature 387, 67–73 (1997).
Chen, S. Y. et al. Global appraisal of the meiotic crossover landscape. Dev. Corpuscle 15, 401–415 (2008).
Rockmill, B., Voelkel-Meiman, K. & Roeder, G. S. Centromere-proximal crossovers are associated with avant-garde break of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174, 1745–1754 (2006).
Vialard, F. et al. Affirmation of a aerial admeasurement of abortive asymmetric break of sister chromatids in the aboriginal arctic bodies of women of avant-garde age. Hum. Reprod. 21, 1172–1178 (2006).
Pellestor, F., Andreo, B., Arnal, F., Humeau, C. & De-maille, J. Affectionate crumbling and chromosomal abnormalities: new abstracts fatigued from in vitro unfertilized animal oocytes. Hum. Genet. 112, 195–203 (2003).
Pellestor, F., Andreo, B., Arnal, F., Humeau, C. & De-maille, J. Mechanisms of non-disjunction in animal changeable meiosis: the co-existence of two modes of malsegregation apparent by the karyotyping of 1,397 in-vitro unfertilized oocytes. Hum. Reprod. 17, 2134–2145 (2002).
Wolstenholme, J. & Angell, R. R. Affectionate age and trisomy — a accumulation apparatus of formation. Chromosoma 109, 435–438 (2000).
Hodges, C. A., Revenkova, E., Jessberger, R., Hassold, T. J. & Hunt, P. A. SMC1beta-deficient changeable mice accommodate affirmation that cohesins are a missing articulation in age-related nondisjunction. Nature Genet. 37, 1351–1355 (2005). This cardboard finds that cohesin-deficient mice appearance meiotic allegory errors that phenocopy age-related errors that are empiric in humans. The after-effects accommodate an arresting adumbration to the account of 'the age effect', in which meiotic missegregation badly increases with affectionate age.
Carlile, T. M. & Amon, A. Meiosis I is accustomed through division-specific translational ascendancy of a cyclin. Corpuscle 133, 280–291 (2008).
Shannon, K. B. & Salmon, E. D. Chromosome dynamics: new ablaze on Aurora B kinase function. Curr. Biol. 12, R458–460 (2002).
Wang, H. W. et al. Architectonics of the Dam1 kinetochore arena circuitous and implications for microtubule-driven accumulation and force-coupling mechanisms. Nature Struct. Mol. Biol. 14, 721–726 (2007).
Cheeseman, I. M. et al. Phospho-regulation of kinetochore–microtubule accessories by the Aurora kinase Ipl1p. Corpuscle 111, 163–172 (2002).
Kang, J. et al. Anatomic cooperation of Dam1, Ipl1, and the close centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Corpuscle Biol. 155, 763–774 (2001).
Li, Y. et al. The mitotic arbor is appropriate for loading of the DASH circuitous assimilate the kinetochore. Genes Dev. 16, 183–197 (2002).
Vogt, E., Kirsch-Volders, M., Parry, J. & Eichenlaub-Ritter, U. Arbor formation, chromosome allegory and the arbor checkpoint in beastly oocytes and susceptibility to meiotic error. Mutat. Res. 651, 14–29 (2008).
Lew, D. J. & Burke, D. J. The arbor accumulation and arbor position checkpoints. Annu. Rev. Genet. 37, 251–282 (2003).
Ke, Y. W., Dou, Z., Zhang, J. & Yao, X. B. Action and adjustment of Aurora/Ipl1p kinase ancestors in corpuscle division. Corpuscle Res. 13, 69–81 (2003).
Courtwright, A. M. & He, X. Dam1 is the appropriate one: phosphoregulation of kinetochore biorientation. Dev. Corpuscle 3, 610–611 (2002).
Terada, Y. Role of chromosomal commuter circuitous in chromosome allegory and cytokinesis. Corpuscle Struct. Funct. 26, 653–657 (2001).
Lengronne, A. et al. Cohesin alteration from sites of chromosomal loading to places of allied transcription. Nature 430, 573–578 (2004).
["618.86"] Mitosis and Meiosis | Where In The Body Does Meiosis Occur
Mitosis and Meiosis | Where In The Body Does Meiosis Occur["552.9"]
 Meiosis - Science | Where In The Body Does Meiosis Occur
Meiosis - Science | Where In The Body Does Meiosis Occur["194"]
 Laurel Murray's Human Biology Site: Compendium Review: Unit 4 ... | Where In The Body Does Meiosis Occur
Laurel Murray's Human Biology Site: Compendium Review: Unit 4 ... | Where In The Body Does Meiosis Occur["1017.53"]